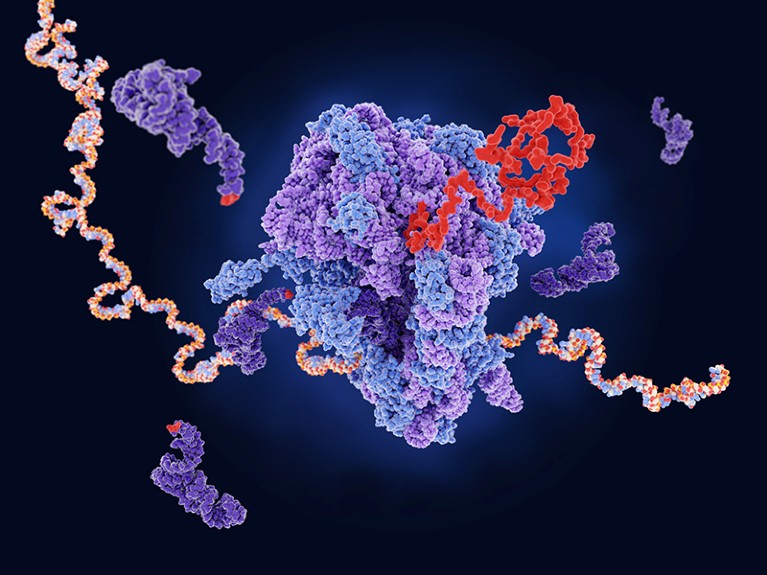
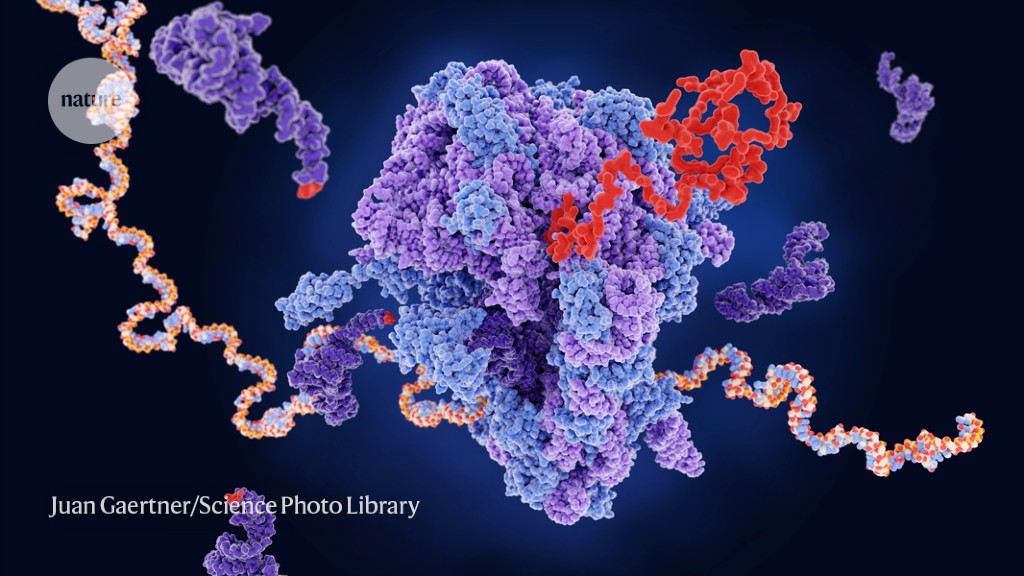
A ribosome (centre) builds a new protein (red) from messenger RNA (multicoloured).Credit: Juan Gaertner/Science Photo Library
For most of the history of life on Earth, genetic information has been carried in a code that specifies just 20 amino acids. Amino acids are the building blocks of proteins, which do most of the heavy lifting in the cell; their side-chains govern protein folding, interactions and chemical activities. By limiting the available side chains, nature effectively restricts the kinds of reaction that proteins can perform.
As a doctoral student in the 1980s, Peter Schultz found himself wondering why nature had restricted itself in this way — and set about trying to circumvent this limitation. Several years later, as a professor at the University of California, Berkeley, Schultz and his team managed to do so by tinkering with the machinery of protein synthesis. Although confined to a test tube, the work marked a key early success in efforts to hack the genetic code.
Since then, many researchers have followed in Schultz’s footsteps, tweaking the cellular apparatus for building proteins both to alter existing macromolecules and to create polymers from entirely new building blocks. The resulting molecules can be used in research and for the development of therapeutics and materials. But it’s been a hard slog, because protein synthesis is a crucial cellular function that cannot easily be changed.
“It’s a super-exciting field,” says Jason Chin, a synthetic and chemical biologist at the MRC Laboratory of Molecular Biology in Cambridge, UK. “Even from the beginning, I think it was clear that we were doing something really special.”
From vitro to vivo
To understand why, we have to consider how proteins are made (see ‘Protein hacking’).
During transcription, DNA instructions are copied into RNA, which is then translated into proteins in a molecular machine called a ribosome. Each triplet of bases in the RNA message (adenine, cytosine, guanine and uracil) represents a word, or codon, of the genetic code. There are 64 words in total — 61 that encode for amino acids and 3 that signal the ribosome to stop.
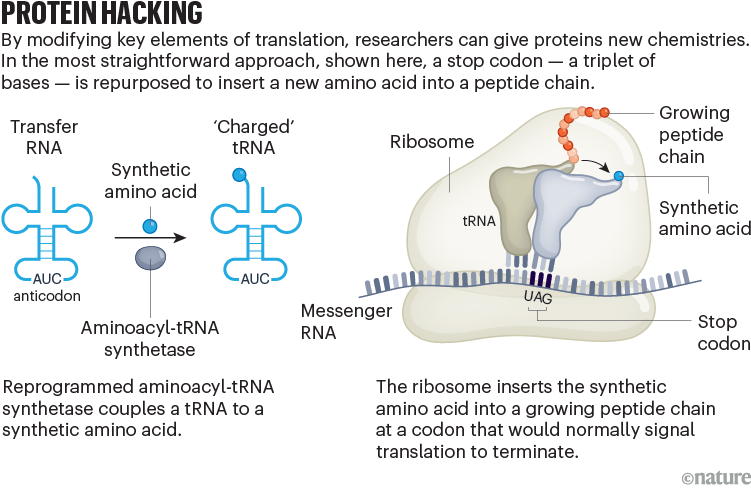
Amino acids are brought to the ribosome by transfer RNAs (tRNAs), each of which corresponds to a complementary codon. Enzymes known as aminoacyl-tRNA synthetases couple the tRNA to its cognate amino acid.
Making this system accept unnatural, or ‘non-canonical’, amino acids in a test tube required several key tweaks. First, Schultz’s team devised a way to chemically attach unnatural amino acids to a tRNA that could recognize one of the three stop codons. They then introduced a stop codon into the gene for the penicillin-resistance protein, β-lactamase. Using these modifications, the researchers explored how different non-canonical protein variants affected enzyme activity1.
But that was in a test tube. To incorporate non-canonical amino acids into living cells, the team had to supplement the translational apparatus without altering it. They did so by identifying and modifying tRNAs and aminoacyl-tRNA synthetases that were bio-orthogonal to the host cell — that is, unable to recognize (and be recognized by) other parts of the cell’s translational machinery.
How to hack the genome
“People thought that would be very, very difficult to do,” says Schultz, who is now the president and chief executive of Scripps Research in La Jolla, California. “Luckily, we were naive enough to think that we could do it.” (Schultz has founded several companies, including Ambrx based in La Jolla, which develops biotherapeutics using non-canonical amino acids. He is currently a member of Ambrx’s scientific advisory board.)
Eventually, Schultz’s team settled on a tRNA-synthetase pair from an archaeon, Methanococcus jannaschii, which did not readily recognize the tRNAs or the synthetases from the Escherichia coli bacteria they planned to modify. The team optimized the molecules to be selective to the new amino acid. In 2001, the researchers engineered this system into E. coli, enabling the cells to incorporate non-standard amino acids into proteins using its own translational machinery2.
“That was probably the beginning of the expansion of the genetic code in vivo,” says Chin, who was formerly a postdoc in Schultz’s laboratory.
A growing toolkit
Schultz’s team and others have used this approach to genetically encode more than 200 non-standard amino acids into proteins, providing a powerful tool with which to study protein structure and function. Scientists could, for instance, introduce fluorescent markers or other labels into proteins, or conduct photocaging experiments, in which proteins are rendered inactive (‘caged’) by chemical groups that can be removed with light.
Scientists have also moved beyond E. coli to hack the genetic codes of nematode worms, fruit flies, plants and even mice. Chin, along with Michael Hastings, a neuroscientist at the University of Cambridge, and their colleagues, for instance, hacked the tRNA and tRNA synthetase for the amino acid pyrrolysine to incorporate an amino acid called alkyne lysine N6-[(2-propynyloxy)carbonyl]-l-lysine (AlkK) into proteins. They used it as a switch to turn genes on and off in mouse brain cells. The researchers deleted a key protein involved in regulating circadian rhythms — daily cycles controlled by the body’s internal clock — and introduced a new version of the gene into the cells that could be translated only in the presence of AlkK. Because the mice couldn’t make that amino acid on their own, the researchers could control their circadian rhythms by adding or removing the amino acids from the rodents’ drinking water3.
Synthetic bacterial genome upgraded for viral defence and biocontainment
For his part, Schultz has been using the expanded genetic code to ask what organisms would look like if they had more than 20 amino acids to play with. In one study, his team created a library of variants of E. coli homoserine O-succinyltransferase, an enzyme involved in the biosynthesis of amino acid methionine that is particularly sensitive to temperature, in which each codon outside the enzyme’s catalytic centre was replaced with the non-canonical amino acid, (p-benzoylphenyl)alanine (pBzF). The team discovered a variant that was stable at up to 21 °C above the typical range — a feat that they attributed to the formation of a chemical bond between pBzF and another amino acid4.
Chin, Schultz and other researchers have also begun to apply this technology to the development of therapeutics. Lei Wang, a chemical biologist at the University of California, San Francisco, for instance, is developing protein-based drugs that can form covalent bonds with other biomolecules.
Proteins typically interact with other molecules through relatively weak, non-covalent interactions, but covalent bonding could enhance their potency, says Wang. In 2020, his team incorporated the non-canonical amino acid fluorosulfate-l-tyrosine (FSY) into PD-1, an immune checkpoint protein that helps to rein in the body’s immune response, to create an anti-tumour drug. Typically, the interaction between PD-1 on T cells, and PD-L1 on tumour cells, dampens the immune response, allowing the tumour to escape immune surveillance. When Wang’s team injected the FSY-containing PD-1 into mice engrafted with human cancer cells, the protein formed an irreversible covalent bond with PD-L1, causing the tumours to shrink5. Wang is a scientific adviser at the company Enlaza Therapeutics in La Jolla, which has developed this therapeutic strategy.
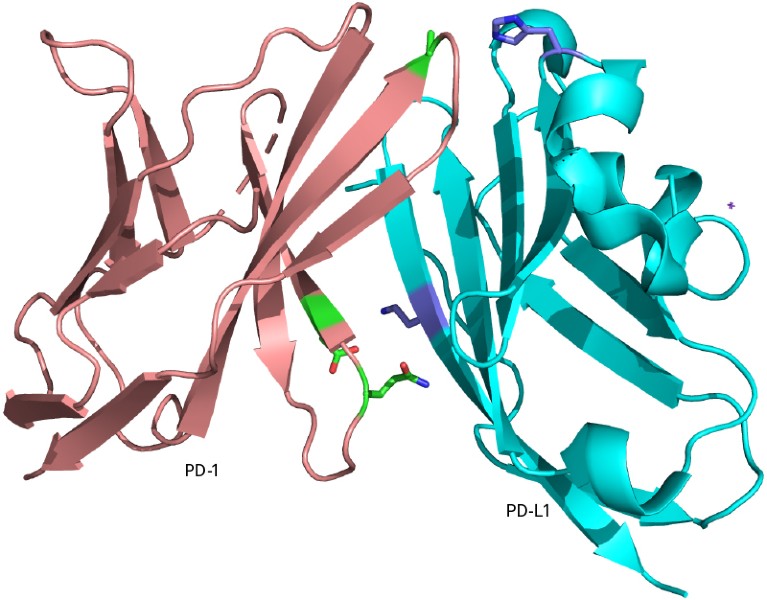
PD-1 (left), an immune checkpoint protein, bound to its ligand PD-L1.Credit: Q. Li et al. Cell 182, 85–97 (2020)
Meanwhile, in Beijing, Tao Liu, a chemical and synthetic biologist at Peking University, and his team have been applying genetic expansion to cell and gene therapies. In a 2021 study, Liu and his team reported engineering cells that, in the presence of the synthetic amino acid O-methyl tyrosine, express insulin6. When implanted into diabetic mice, the researchers could control the animal’s blood glucose levels by controlling how much O-methyl tyrosine they dispensed in the animals’ food.
Expanding applications
Beyond encoding new amino acids, one application of an extended genetic code is genetic isolation. With 61 codons for 20 amino acids, the genetic code is redundant, meaning that multiple codons encode the same amino acid. It is also nearly universal. By replacing all instances of a given codon for a synonymous one and removing the machinery that uses the original codon, researchers can render the cell effectively immune to foreign DNA — including pathogens.
In a 2022 study Chin and his group demonstrated this concept; they created an E. coli mutant in which two of its six serine codons were reassigned to encode for other amino acids — then they deleted the tRNAs that recognized the original serine codons. They further tweaked the system to ensure that viruses couldn’t use their own tRNAs, if they had them. The resulting cells were resistant to horizontal gene transfer from other bacteria, as well as to viral infection7. Another group, led by synthetic biologist George Church at Harvard University in Cambridge, Massachusetts, applied a similar ‘genetic firewall’ approach to create phage-resistant bacteria in a paper published earlier this year8.
Researchers can also use a modified genetic code to create polymers. In 2021, Chin’s team hacked the genetic code to synthesize short polymers, and even an artificial circular structure called a macrocycle, in E. coli9. Now, Chin hopes to push this technology further to create cell factories that can churn out entirely new polymers, such as plastics. Like proteins, plastics are composed of long strings of monomers. But, although the genetic code dictates the sequence of amino acids in proteins, no equivalent system exists for artificial polymers.
“If you could encode the expanded set of building blocks in the same way that we can proteins, then we could turn cells into living factories for the encoded synthesis of polymers for everything from new drugs to materials,” says Chin. (Chin founded a company, Constructive Bio based in Saffron Walden, UK, to advance this goal.)
Pushing boundaries
Besides codon reassignment, researchers can also increase the number of available protein building blocks by expanding the nucleic acid alphabet from four bases to six, thus increasing the number of possible triplet codons to 216. In 2014, a team led by biochemist Floyd Romesberg, who was then at Scripps Research, reported creating a bacterial strain with a six-base genetic alphabet that could successfully replicate10. The group subsequently demonstrated that these cells could use their expanded DNA to produce proteins containing non-canonical amino acids11.
NatureTech hub
Another approach is to expand the length of a codon from three bases to four, thus increasing the number of possible codons to 256. This requires modifying multiple pieces of the translation machinery, including the ribosome. Chin and his team have used this strategy to incorporate four non-canonical amino acids into E. coli on a transcript that also contains conventional three-base codons12. Other researchers have been exploring whether it might even be possible to create an all-quadruplet genetic code.
Some researchers are attempting more extreme alterations. These include backbone modifications — creating so-called β- or γ-amino-acids (as opposed to α-amino-acids, which are found in nature), or amino acids that are reverse mirror images of standard amino acids. Polymers built from either type of building block would probably be highly stable, because typical protein-degrading machinery would not be able to recognize them.
But existing translational machinery isn’t built to accept these exotic amino acids, including the aminoacyl-tRNA synthetases that attach amino acids onto tRNA. Hiroaki Suga, a chemical biologist at the University of Tokyo, developed one workaround in 2006 with the ‘flexizyme’, an RNA-based catalyst that can perform the job of protein synthetases: linking amino acids to tRNAs13.
Suga focuses on synthesizing polymers in vitro, because it provides the greatest flexibility in terms of the modifications he can make. Using the flexizyme, his team combined up to 11 non-canonical amino acids with 12 standard amino acids in a single macrocycle14. Suga co-founded the biotechnology company PeptiDream, based in Kawasaki, Japan, to develop drugs using the same technology.
Again, however, that work is in vitro; applying backbone- or configuration-modified amino acids to proteins in cells remain a challenge. Although there are a few instances in which scientists have been able to incorporate these more exotic amino acids, further steps are necessary to make the process more efficient. As well as finding synthetases that could work with these amino acids in vivo, in many cases scientists would have to design a ribosome that can process these new amino acids — while still carrying out its usual duties.
In two recent studies Alanna Schepartz, a chemical and synthetic biologist at the University of California, Berkeley, and her team report steps toward solving these problems. In one of the papers, they describe a synthetase from the archaeon Methanomethylophilus alvus that can accept non-α-amino-acids and is bio-orthogonal to E. coli. In the other study, the team reports a computational technique for screening unique backbone monomers that the E. coli ribosome can process efficiently. The researchers say this will help to identify non-α-amino-acids that are most likely to be compatible with an existing ribosome15,16.
Other groups are working to develop a ribosome that can produce proteins with exotic amino acids from scratch. Last October, researchers at Westlake University in Hangzhou, China, reported a mirror-image RNA polymerase that could synthesize all the RNA molecules that are needed to produce a mirror-image ribosome17. Although many more steps are needed to make a mirror-image ribosome a reality, creating such a molecule would be an important step towards using the translational machinery to make mirror-image proteins.
Other researchers are working to reprogram the ribosome to create polymers with carbon–carbon bonds, as opposed to the nitrogen–carbon amide bonds they typically forge to link amino acids.
Should these strategies come to fruition, they would endow scientists with tremendous synthetic power over new polymers. Nobody knows what properties can emerge from a synthetic polymer that’s created with the same length and level of sequence definition as a protein, “because no such molecules have ever been made”, Schepartz says. But she and others hope to get there soon. “It’s a very exciting time for the field.”






More News
How I fled bombed Aleppo to continue my career in science
Powerful ‘nanopore’ DNA sequencing method tackles proteins too
US funders to tighten oversight of controversial ‘gain-of-function’ research