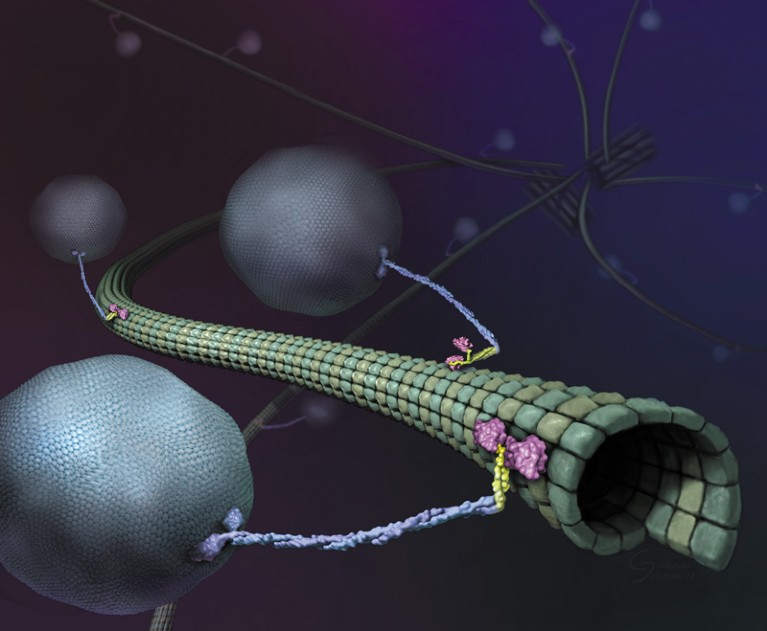
Illustration of molecular motors called kinesins on a microtubule.Credit: Graham Johnson, Ron Vale/HHMI
Almost as soon as there were super-resolution microscopes, scientists pointed them towards molecular motors called kinesins. These proteins, powered by the molecular fuel ATP, drive crucial processes including cell division, cell signalling and intracellular transport by shuttling cargo along protein highways called microtubules. Researchers have long wanted to understand how these motors work, but to visualize them, scientists have had to slow them down or isolate them in simplified, in vitro systems.
Now, in papers published concurrently in Science, two teams working independently have used a super-resolution tool called MINFLUX to study the motor in near-real time at physiologically relevant concentrations of ATP. The first paper, led by MINFLUX’s inventor, Stefan Hell, who has a joint appointment at the Max Planck Institute (MPI) for Multidisciplinary Sciences in Göttingen and the MPI for Medical Research in Heidelberg, both in Germany, used a new instrument design to track the protein in 3D, revealing details about its motion1. The second, led by biophysicist Jonas Ries at the European Molecular Biology Laboratory in Heidelberg, showed for the first time that MINFLUX is capable of tracking kinesin even amid the bustle of living cells2.
“This technology requires a lot of different things to work, and it’s fun to see all of these things coming together,” says Michelle Digman, a biomedical engineer at the University of California, Irvine, who develops imaging strategies but was not involved in either study. “It seemed like a proof of concept to show that they’re able to track kinesin very precisely. And when you have the live cell system, that’s even more spectacular.”
Walking the walk
Researchers began working out the basics of kinesin’s motion shortly after the protein’s discovery in 1985, but the advent of super-resolution tools brought a new level of detail. In 2004, researchers used a technique called FIONA (fluorescence imaging with one-nanometre accuracy) to show that kinesin, which looks like a tall, twisted stalk wearing oversized shoes, walks ‘hand-over-hand’ on its microtubule track, moving its feet in a motion similar to a child’s hands as they traverse a set of monkey bars3. But, although FIONA provided exceptional spatial resolution, the scientists had to ration ATP to slow the protein down enough to study it. In the past decade, researchers tagged kinesin with beads of germanium4 or gold5 to track it, but these relatively bulky tags leave doubt over how well the methods recapitulate the protein’s full range of motion.
When Hell and his team introduced6 MINFLUX in 2016, he saw it as an advance over its predecessor, stimulated emission depletion (STED) microscopy, for which Hell shared the 2014 Nobel Prize in Chemistry. STED uses a doughnut-shaped ‘depletion’ laser superimposed on an excitation laser to effectively shrink the area of fluorescence below the conventional diffraction limit of light (about 250 nanometres). MINFLUX, by contrast, uses a doughnut-shaped laser to create a point of zero fluorescence intensity at its centre. By moving this laser, researchers can pinpoint a fluorescent molecule’s location at near-physiological speeds.
Smart microscopes spot fleeting biology
In the new study1, published in March, Hell’s group tested a version of MINFLUX that pulses linear lasers in two directions in the focal plane in quick succession, localizing the protein by finding where the overlapping fluorescence intensities are lowest. By combining multiple measurements, the researchers were able to produce tracks that show where the molecule is moving along the microtubule, like an app that maps out a runner’s path.
Although the new MINFLUX wasn’t much more effective than the previous iteration, Hell says, his team was able to use it to deduce a walking speed for kinesin of 550 nanometres per second and a stride length of 16 nanometres at ATP concentrations similar to those found in living cells. By tagging and tracking different parts of the protein, the team also showed that each stride comprises two 8-nanometre substeps, and that a kinesin’s stalk rotates as it moves, resulting in a forward motion that twirls slightly to the right. The authors also found that ATP is taken up when only one foot is bound to the microtubule, but is consumed when both feet are bound — resolving previously contradictory findings7,8.
Ries and his team approached kinesin motion from a different perspective. They used a commercial MINFLUX instrument developed by Abberior, a Göttingen-based company co-founded by Hell, to track the protein in living cells. The crowded and ever-changing cellular environment meant that the group ended up with fewer tracks, but they were able to capture side-stepping, stalling and hops from one microtubule to another — all efforts by the protein to circumvent roadblocks that aren’t typically seen in purified samples. “Otherwise, we see very much the same things: similar walking speeds, the same step sizes,” Ries says. “And it was nice to be able to measure those for the first time in the living cell.”
William Hancock, a biomedical engineer at Pennsylvania State University in State College who studies kinesin but was not involved in either study, calls the studies “pretty incredible”. The results, he notes, largely gel with past work, almost all of which had been done in vitro using purified motor proteins. “We get our very specific answers there, but it’s nice to be able to extrapolate back to the inside of cells. It’s really a tour de force.”
Motoring forwards
That said, it’s clear to Hell that MINFLUX “is not yet at the limit of its performance”. Having achieved subnanometre spatial resolution, one area where MINFLUX can still improve, he says, is in its temporal performance. Both groups worked with ATP concentrations of up to one millimolar, but the average cell contains more than four times as much.
Achieving real-time temporal resolution under those conditions will require better tracking systems, says Luke Lavis, an organic chemist at the Howard Hughes Medical Institute’s Janelia Research Campus in Ashburn, Virginia, who developed the dyes used in Ries’s study. “This revolution in super-resolution imaging brings into focus, if I can say that, the fact that a lot of the labelling and attachment strategies we’ve used are big,” he says. “The dream for all of us is being able to attach a small-molecule fluorophore to a specific place on a protein where the linker is just a couple of atoms.” New tools such as genetic code expansion, in which synthetic amino acids are engineered into proteins to facilitate labelling, are moving in this direction, he adds, but “they’re far from being turn-key”.
NatureTech hub
Such advances could open multiple research avenues, especially if they allow researchers to track several proteins, or several sites in proteins, at once. This year, researchers introduced a tool called RESI (resolution enhancement by sequential imaging), which aims to do just that9. RESI can label adjacent copies of the same target molecule with different tags, allowing scientists to distinguish between molecules that are less than one nanometre apart. Although RESI currently works only on fixed or stationary molecules, combining its findings with MINFLUX tracking data on non-fixed copies could yield complementary findings about a protein’s arrangement and motion.
Ries is interested in studying other motor proteins, and in a supplementary experiment shared in his paper2, applied MINFLUX to the muscle-contracting protein, myosin. In 2019, Lavis co-founded a company to use single molecule tracking for drug development. Other scientists have suggested everything from transcription factors and DNA-binding proteins to cohesin and other molecular motors as targets for future study.
But Haley Marks, a biomedical engineer and project scientist at the California NanoSystems Institute at the University of California, Los Angeles, says it will take time before MINFLUX becomes a truly accessible and affordable “plug-and-play” system. In the meantime, she suggests, MINFLUX could benefit the wider community by contributing to virtual super-resolution tools. These artificial-intelligence algorithms train on large collections of microscopy images to learn how to create super-resolution images from conventional microscopy ones. The resulting data, Marks says, “are much more accessible by people who can’t afford a multimillion-dollar microscope”.







More News
Author Correction: Stepwise activation of a metabotropic glutamate receptor – Nature
Changing rainforest to plantations shifts tropical food webs
Streamlined skull helps foxes take a nosedive